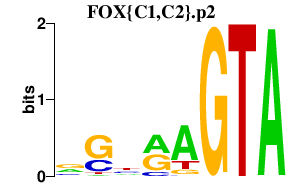
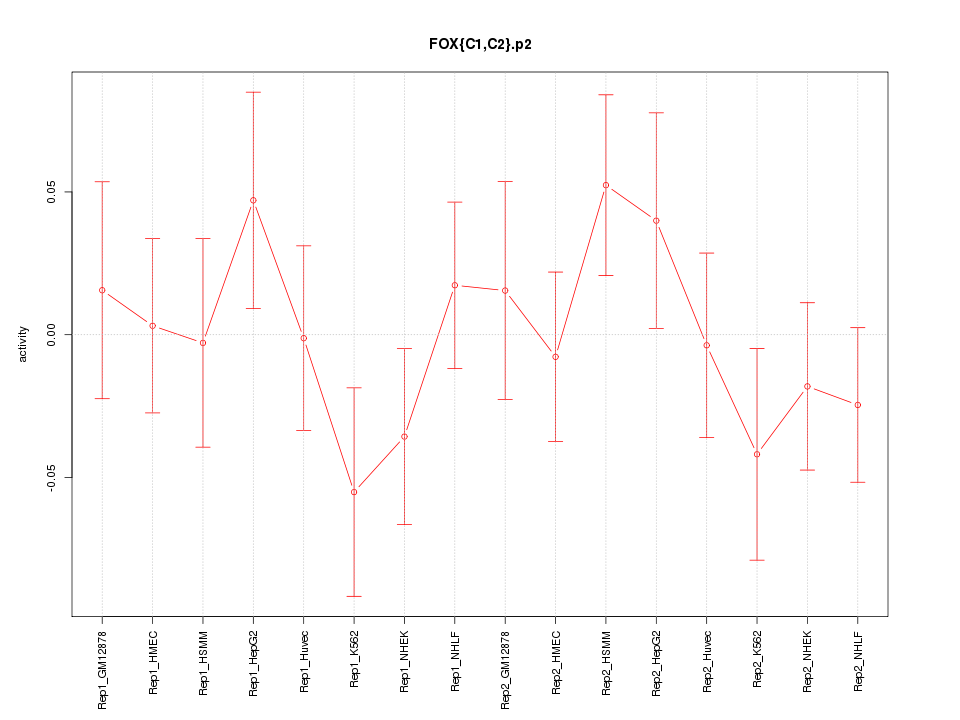
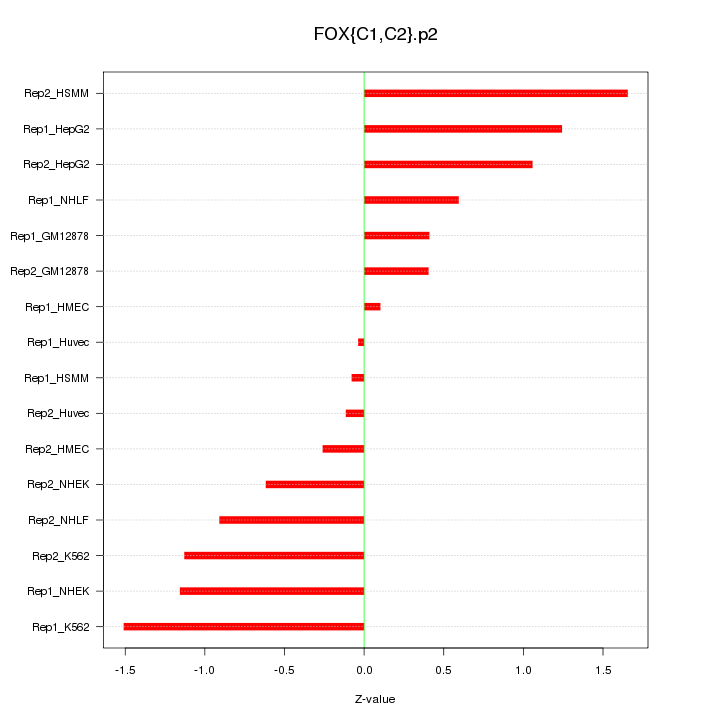
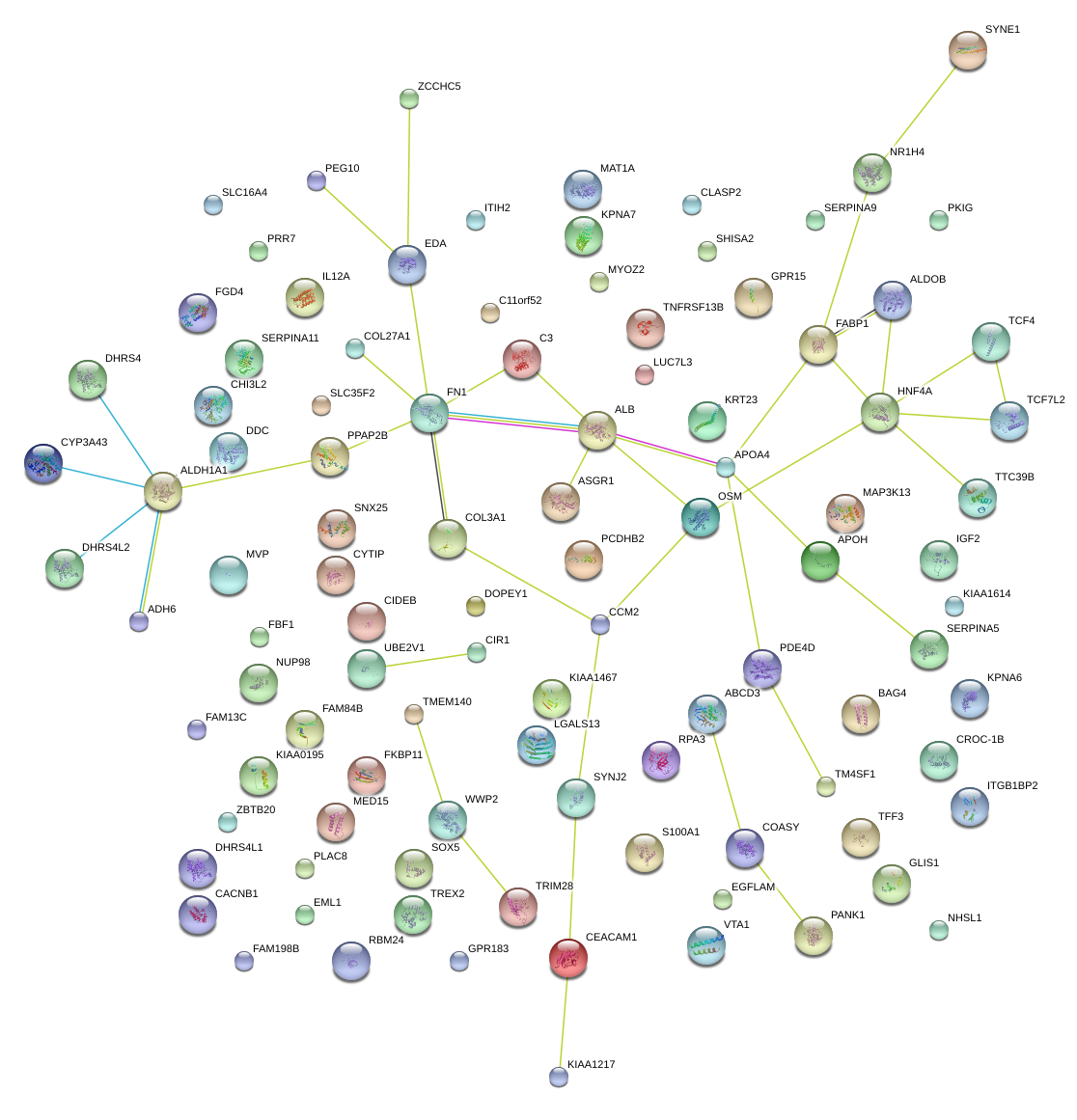
|
chr3_-_115826481
|
2.306
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr17_-_61655915
|
1.596
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr4_+_74491272
|
1.548
|
|
ALB
|
albumin
|
|
chr14_-_93988838
|
1.318
|
NM_001080451
|
SERPINA11
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11
|
|
chr10_+_7785347
|
1.144
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr10_+_7785360
|
1.064
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr2_-_216008689
|
1.030
|
|
FN1
|
fibronectin 1
|
|
chr10_+_7785331
|
1.012
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr10_+_7785290
|
0.995
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr14_+_94117511
|
0.986
|
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr4_+_186368277
|
0.959
|
NM_031953
|
SNX25
|
sorting nexin 25
|
|
chr3_-_150578023
|
0.956
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr4_+_74500748
|
0.885
|
|
ALB
|
albumin
|
|
chr3_-_150578103
|
0.850
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr14_+_94117483
|
0.835
|
NM_000624
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr12_+_99391870
|
0.833
|
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr10_+_7785235
|
0.817
|
NM_002216
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr4_-_100359353
|
0.806
|
NM_000672
NM_001102470
|
ADH6
|
alcohol dehydrogenase 6 (class V)
|
|
chr6_+_158358067
|
0.795
|
NM_001178088
|
SYNJ2
|
synaptojanin 2
|
|
chr1_+_111573855
|
0.786
|
NM_001025199
|
CHI3L2
|
chitinase 3-like 2
|
|
chr17_-_7023378
|
0.780
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr3_+_186529376
|
0.776
|
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr12_+_99391681
|
0.765
|
NM_005123
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr22_+_19239221
|
0.727
|
|
MED15
|
mediator complex subunit 15
|
|
chr6_-_152744022
|
0.723
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr2_+_189547358
|
0.708
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189547377
|
0.682
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr20_+_42417752
|
0.680
|
NM_001030003
NM_001030004
NM_175914
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr2_+_189547323
|
0.674
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_-_88208650
|
0.656
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr17_-_7023283
|
0.656
|
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr7_+_134483305
|
0.644
|
NM_018295
|
TMEM140
|
transmembrane protein 140
|
|
chr3_-_150578148
|
0.615
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr17_+_70984173
|
0.611
|
|
KIAA0195
|
KIAA0195
|
|
chr4_+_120276386
|
0.609
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr9_+_115958051
|
0.599
|
NM_032888
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr7_+_99263571
|
0.569
|
NM_022820
NM_057095
NM_057096
|
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43
|
|
chr22_-_28992787
|
0.563
|
NM_020530
|
OSM
|
oncostatin M
|
|
chr5_+_176806401
|
0.562
|
NM_001174101
NM_030567
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr17_-_16816113
|
0.535
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr4_-_159312973
|
0.503
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr14_+_99309688
|
0.501
|
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr2_-_158008763
|
0.495
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr11_-_116199145
|
0.489
|
NM_000482
|
APOA4
|
apolipoprotein A-IV
|
|
chr10_-_91393610
|
0.477
|
NM_138316
NM_148978
|
PANK1
|
pantothenate kinase 1
|
|
chr11_-_2113297
|
0.472
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr10_+_24538094
|
0.457
|
|
KIAA1217
|
KIAA1217
|
|
chr21_-_42608531
|
0.453
|
|
TFF3
|
trefoil factor 3 (intestinal)
|
|
chr19_-_6671585
|
0.439
|
NM_000064
|
C3
|
complement component 3
|
|
chr9_-_103237851
|
0.424
|
NM_000035
|
ALDOB
|
aldolase B, fructose-bisphosphate
|
|
chr9_-_74735669
|
0.424
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr4_-_159313167
|
0.422
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr14_+_94117545
|
0.411
|
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr5_+_38481392
|
0.396
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr3_-_116272911
|
0.393
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr10_-_82039158
|
0.381
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr13_-_98757634
|
0.353
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr9_-_15240113
|
0.347
|
NM_001168342
|
TTC39B
|
tetratricopeptide repeat domain 39B
|
|
chr12_-_23995220
|
0.347
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr3_+_99733432
|
0.340
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr16_+_68516401
|
0.340
|
NM_199424
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr14_+_23527866
|
0.340
|
NM_198083
|
DHRS4
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4
dehydrogenase/reductase (SDR family) member 4 like 2
|
|
chrX_+_70438352
|
0.329
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chr4_-_84254932
|
0.329
|
NM_001130715
NM_016619
|
PLAC8
|
placenta-specific 8
|
|
chr5_+_140454382
|
0.328
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr4_-_84254912
|
0.324
|
|
PLAC8
|
placenta-specific 8
|
|
chr7_-_50600536
|
0.319
|
NM_001082971
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr16_+_29739256
|
0.316
|
|
MVP
|
major vault protein
|
|
chr9_+_674193
|
0.311
|
|
|
|
|
chr6_+_142510034
|
0.305
|
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr20_+_42644633
|
0.304
|
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chr6_-_138862271
|
0.301
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr16_+_29739215
|
0.299
|
NM_005115
NM_017458
|
MVP
|
major vault protein
|
|
chr13_-_25523164
|
0.299
|
NM_001007538
|
SHISA2
|
shisa homolog 2 (Xenopus laevis)
|
|
chr7_-_98643024
|
0.299
|
NM_001145715
|
KPNA7
|
karyopherin alpha 7 (importin alpha 8)
|
|
chr19_-_47723183
|
0.298
|
|
CEACAM1
|
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein)
|
|
chr17_-_36347197
|
0.296
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr1_+_179148935
|
0.292
|
NM_020950
|
KIAA1614
|
KIAA1614
|
|
chr4_-_84254873
|
0.291
|
|
PLAC8
|
placenta-specific 8
|
|
chr3_-_33675484
|
0.288
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr11_+_111294868
|
0.288
|
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr12_+_32546243
|
0.278
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr2_-_174968595
|
0.277
|
NM_004882
|
CIR1
|
corepressor interacting with RBPJ, 1
|
|
chr17_-_45620313
|
0.276
|
|
|
|
|
chr17_+_37967690
|
0.274
|
|
COASY
|
CoA synthase
|
|
chr1_+_151867483
|
0.270
|
NM_006271
|
S100A1
|
S100 calcium binding protein A1
|
|
chrX_+_68752635
|
0.267
|
NM_001005609
NM_001005610
NM_001005612
NM_001005613
NM_001399
|
EDA
|
ectodysplasin A
|
|
chr1_-_53972464
|
0.265
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr5_-_59819642
|
0.261
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_+_46176105
|
0.260
|
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr1_-_56750235
|
0.258
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr11_-_3759886
|
0.255
|
|
NUP98
|
nucleoporin 98kDa
|
|
chr7_+_94123672
|
0.255
|
|
PEG10
|
paternally expressed 10
|
|
chr11_+_111294810
|
0.254
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr1_-_110735158
|
0.244
|
NM_004696
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chr18_-_51219880
|
0.244
|
|
TCF4
|
transcription factor 4
|
|
chr17_+_37967702
|
0.242
|
|
COASY
|
CoA synthase
|
|
chr6_+_142510075
|
0.242
|
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr10_-_112668892
|
0.240
|
NM_001195304
NM_001195305
NM_001195307
|
BBIP1
|
BBSome interacting protein 1
|
|
chr17_+_37967617
|
0.240
|
NM_001042529
NM_001042530
NM_001042531
NM_001042532
NM_025233
|
COASY
|
CoA synthase
|
|
chr14_-_23846904
|
0.239
|
|
CIDEB
|
cell death-inducing DFFA-like effector b
|
|
chr3_+_161189322
|
0.238
|
NM_000882
|
IL12A
|
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35)
|
|
chrX_-_77801480
|
0.238
|
NM_152694
|
ZCCHC5
|
zinc finger, CCHC domain containing 5
|
|
chr7_-_7724762
|
0.238
|
NM_002947
|
RPA3
|
replication protein A3, 14kDa
|
|
chr6_+_17390910
|
0.233
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr1_+_94656517
|
0.233
|
NM_001122674
NM_002858
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3
|
|
chr10_-_60792225
|
0.228
|
NM_001001971
NM_001166698
NM_198215
|
FAM13C
|
family with sequence similarity 13, member C
|
|
chr6_+_83834080
|
0.227
|
NM_015018
|
DOPEY1
|
dopey family member 1
|
|
chr17_+_7402315
|
0.224
|
NM_001198622
NM_001198623
NM_001198624
NM_003808
NM_172087
NM_172088
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr10_+_60607232
|
0.223
|
NM_001143774
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like
|
|
chr17_-_45626364
|
0.222
|
|
|
|
|
chr11_-_85107569
|
0.218
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr1_-_36721010
|
0.213
|
|
CSF3R
|
colony stimulating factor 3 receptor (granulocyte)
|
|
chr6_-_138862273
|
0.212
|
|
|
|
|
chr16_+_85158357
|
0.212
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr7_+_143263191
|
0.211
|
NM_001004685
|
OR2F2
|
olfactory receptor, family 2, subfamily F, member 2
|
|
chr6_+_142510108
|
0.209
|
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr22_+_38246509
|
0.207
|
NM_001675
NM_182810
|
ATF4
|
activating transcription factor 4 (tax-responsive enhancer element B67)
|
|
chr11_+_111288686
|
0.207
|
|
HSPB2-C11ORF52
HSPB2
|
HSPB2-C11orf52 read-through transcript
heat shock 27kDa protein 2
|
|
chr6_+_12825818
|
0.205
|
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr11_-_8789457
|
0.203
|
NM_139157
|
ST5
|
suppression of tumorigenicity 5
|
|
chr2_-_190635566
|
0.203
|
NM_005259
|
MSTN
|
myostatin
|
|
chr14_-_93924663
|
0.202
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr4_+_146820805
|
0.201
|
NM_001080531
|
C4orf51
|
chromosome 4 open reading frame 51
|
|
chr2_-_215998181
|
0.201
|
|
FN1
|
fibronectin 1
|
|
chr5_-_35266305
|
0.201
|
|
PRLR
|
prolactin receptor
|
|
chr9_-_130683997
|
0.197
|
NM_001122671
NM_001122672
NM_004059
|
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic
|
|
chr12_+_6424293
|
0.197
|
NM_001242
|
CD27
|
CD27 molecule
|
|
chr10_+_86174665
|
0.194
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr13_-_19244398
|
0.194
|
|
PSPC1
|
paraspeckle component 1
|
|
chr6_+_142510094
|
0.193
|
NM_016485
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr3_-_42892195
|
0.192
|
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chr14_-_93924873
|
0.191
|
NM_000295
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr1_-_25164061
|
0.188
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr14_+_87541220
|
0.187
|
NM_003608
|
GPR65
|
G protein-coupled receptor 65
|
|
chr14_+_64041257
|
0.187
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr1_-_24112345
|
0.185
|
NM_001841
|
CNR2
|
cannabinoid receptor 2 (macrophage)
|
|
chr6_-_131333173
|
0.185
|
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr9_-_14900992
|
0.184
|
NM_144966
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr2_+_149895274
|
0.184
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chrX_-_130250880
|
0.183
|
|
IGSF1
|
immunoglobulin superfamily, member 1
|
|
chr8_-_110689396
|
0.183
|
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr5_+_140871107
|
0.183
|
|
|
|
|
chr17_-_3546047
|
0.182
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr3_-_13032130
|
0.182
|
|
|
|
|
chr2_+_190710730
|
0.181
|
NM_001042521
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr9_-_94338072
|
0.181
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr17_-_33179118
|
0.180
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr3_-_150578207
|
0.180
|
NM_014220
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr17_-_45626967
|
0.176
|
|
|
|
|
chr1_-_149247464
|
0.176
|
NM_001163258
NM_018379
|
FAM63A
|
family with sequence similarity 63, member A
|
|
chr12_+_15017222
|
0.176
|
NM_006205
|
PDE6H
|
phosphodiesterase 6H, cGMP-specific, cone, gamma
|
|
chr5_-_156468571
|
0.171
|
|
HAVCR2
|
hepatitis A virus cellular receptor 2
|
|
chr2_-_188127294
|
0.171
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr2_+_33555182
|
0.168
|
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr2_+_158666627
|
0.167
|
NM_173355
|
UPP2
|
uridine phosphorylase 2
|
|
chr17_-_36346240
|
0.167
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr5_+_35892747
|
0.166
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr17_-_6495566
|
0.164
|
NM_016060
|
MED31
|
mediator complex subunit 31
|
|
chr18_-_63333487
|
0.163
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr2_-_202271598
|
0.162
|
NM_033066
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4)
|
|
chr1_+_200246370
|
0.162
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr11_+_117680504
|
0.161
|
NM_000733
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex)
|
|
chr9_+_139265533
|
0.159
|
NM_001004353
|
C9orf173
|
chromosome 9 open reading frame 173
|
|
chr16_-_52295214
|
0.156
|
NM_001127897
NM_015272
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr7_-_50596238
|
0.156
|
NM_000790
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr11_+_111288669
|
0.156
|
NM_001541
|
HSPB2-C11ORF52
HSPB2
|
HSPB2-C11orf52 read-through transcript
heat shock 27kDa protein 2
|
|
chr12_-_44919818
|
0.155
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr13_+_51334117
|
0.152
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr4_+_166519392
|
0.152
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chr11_-_61823078
|
0.152
|
NM_206998
|
SCGB1D4
|
secretoglobin, family 1D, member 4
|
|
chr19_+_40895669
|
0.151
|
NM_014383
|
ZBTB32
|
zinc finger and BTB domain containing 32
|
|
chr2_-_70374349
|
0.151
|
NM_003096
|
SNRPG
|
small nuclear ribonucleoprotein polypeptide G
|
|
chr14_-_53977785
|
0.150
|
NM_005776
|
CNIH
|
cornichon homolog (Drosophila)
|
|
chr2_+_172530120
|
0.150
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr12_+_54510903
|
0.146
|
|
LOC440104
|
1110012D08Rik pseudogene
|
|
chr8_+_11388918
|
0.143
|
NM_001715
|
BLK
|
B lymphoid tyrosine kinase
|
|
chr1_+_94656556
|
0.141
|
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3
|
|
chr15_+_38843535
|
0.140
|
NM_005258
|
GCHFR
|
GTP cyclohydrolase I feedback regulator
|
|
chr2_+_132196533
|
0.140
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chr12_+_4300607
|
0.139
|
NM_020375
|
C12orf5
|
chromosome 12 open reading frame 5
|
|
chr2_+_170074457
|
0.139
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr2_+_69055208
|
0.138
|
NM_019617
|
GKN1
|
gastrokine 1
|
|
chr12_-_110378240
|
0.137
|
|
|
|
|
chr12_+_58276114
|
0.136
|
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr3_+_153499883
|
0.135
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr2_-_233349469
|
0.134
|
NM_001172416
NM_001172417
NM_002242
|
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13
|
|
chr4_+_2853434
|
0.134
|
|
ADD1
|
adducin 1 (alpha)
|
|
chr2_-_70374284
|
0.133
|
|
SNRPG
|
small nuclear ribonucleoprotein polypeptide G
|
|
chr1_-_155788775
|
0.133
|
NM_001195388
NM_031281
|
FCRL5
|
Fc receptor-like 5
|
|
chr13_+_96592051
|
0.131
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr5_-_111121150
|
0.130
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr8_-_124818754
|
0.130
|
NM_001003954
NM_004306
|
ANXA13
|
annexin A13
|
|
chr20_+_56903595
|
0.127
|
|
GNAS
|
GNAS complex locus
|
|
chr14_+_22059212
|
0.126
|
|
|
|
|
chr20_+_56904042
|
0.125
|
|
GNAS
|
GNAS complex locus
|
|
chr10_-_98315090
|
0.124
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr15_+_76171981
|
0.123
|
NM_001101404
|
SH2D7
|
SH2 domain containing 7
|
|
chr1_-_158521491
|
0.122
|
|
PEX19
|
peroxisomal biogenesis factor 19
|
|
chr6_-_152744143
|
0.122
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|